Multivariate analysis of RNA levels from postmortem human brains as measured by three different methods of RT-PCR
Nancy L. Johnstona, Juraj Cerevnakb, Andrew D. Shorec, E. Fuller Torreyb, Robert H. Yolkena*, The Stanley Neuropathology Consortium
aThe Stanley Foundation Neurovirology Laboratory, Department of Pediatrics, Johns Hopkins University, 600 N. Wolfe Street, Baltimore, MD 21287-4933, USA
bNational Institutes of Mental Health, St. Elizabeth’s Hospital, Washington DC 20032, USA
cHealth Services Research and Development Center, The Johns Hopkins School of Hygiene and Public Health, Johns Hopkins University, Baltimore, MD 21215, USA
Abstract
The analysis of RNA from postmortem human brain tissue by reverse transcription-polymerase chain reaction (RT-PCR) provides a practical method to measure both normal and abnormal brain gene expression. A major limitation in using human material is that yields can vary dramatically from individual to individual, making comparisons between sample difficult. In this report, we study the association of pH and several pre- and postmortem factors on the RNA yields from 89 postmortem human occipital cortices. Glyceraldehyde phosphate dehydrogenase (GAPdH) mRNA levels were measured by RT-PCR. A major variant in this method is the priming used in the reverse transcription reaction. Three different methods of reverse transcription were performed and the resultant levels of products compared against the pre- and postmortem factors and pH. The levels of GAPdH correlated significantly to pH and pH itself to the rapidity of death (RoD) (agonal state) indicting that premortem factors may play the greatest role in determining postmortem RNA levels. The three methods of priming showed different sensitivities most notably that oligo dT priming alone is vulnerable to long freezer intervals (FI). We conclude that premortem factors are the major affectors of RNA levels variation and that the poly A tail region of the molecule appears to be adversely affected by extended freezer storage. ©1997 Elsevier Science B.V.
Keywords: Postmortem human brain; pH; RNA; RT-PCR; Psychiatric disorders; Glyceraldehyde phosphate dehydrogenase
1. Introduction
The rapid development of neurosciences in recent years has generated an increasing interest in using human postmortem brain tissue. Such tissue is useful to study normal brain structure and function as well as to identify abnormalities in various neurological and psychiatric disorders. Many such studies rely on the use of RNA to study genes, viruses, neurotransmission or other aspects of cerebral function using techniques such as RT-PCR, Northern blots, in situ hybridization and in vitro translation.
Despite the known relative stability of RNA in postmortem tissue (Perrett et al., 1988; Barton et al., 1993), studies of human postmortem brain tissue often yield RNA levels that vary widely among samples. In contrast to animal studies, in which both pre- and postmortem conditions can be rigidly controlled, the circumstances of life, death and postmortem conditions in humans often differ markedly between cases. Each individual is affected by a unique combination of variables, including the mode of death, the temperature and interval between death and refrigeration of the body–refrigeration interval (RI)–or removal of the brain tissue and freezing or fixing it–postmortem interval (PMI)–and the interval of storage before the tissue is used for research purposes–in the case of frozen tissue, freezer interval (FI)–see Fig. 1.
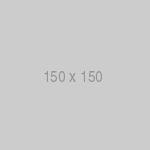
Fig. 1. Handling of body and tissue used in study, RoD, rapidity of death; RI, refrigeration interval; PMI, postmortem interval; FI, freezer interval
Possibly the most significant and most studied premortem factor is the rapidity of death (RoD) or agonal state. Previous research has shown that both RNA and protein levels are affected by the agonal state (Octave et al., 1988; Butterworth et al., 1983; Spokes, 1979; Bowen et al., 1976; Czudek and Reynolds, 1990; Perry et al., 1981; Butterworth, 1986) and that terminal states, such as infections, affect postmortem RNA yield (Taylor et al., 1986). At least three studies have shown that RNA and enzymes deteriorate more rapidly during prolonged agonal states compared to rapid agonal states (Harrison et al., 1991; Dodd et al., 1988; Morrison et al., 1987). Prolonged agonal states and hypoxia produce an increase in tissue lactic acid, which lowers pH (Dodd et al, 1988; Hardy et al., 1985). Measurement of pH has been shown to correlate with RNA levels (Harrison et al, 1995; Kingsbury et al., 1995) and has therefore been used for assessing the agonal state and predicting RNA levels.
Other premortem variations are inevitable, as human samples fall into a heterogeneous group of ages, gender and various diagnostic groups, to name just a few of the attributes. All of these premortem factors may influence individual RNA levels, but no studies have been done to assess their effect on overall RNA yields. Because every individual is a composite of many different factors, variations in brain RNA levels are necessarily going to be the net result of an exceedingly complex set of underlying conditions.
Current methods of RNA detection include direct detection by Northern blots and in situ hybridization or indirect detection by reverse transcription-polymerase chain reaction (RT-PCR). PCR based methods are becoming widely used due to their relative simplicity and the ability of researchers to analyze large sample sets that would be difficult with Northerns and impossible with in situs. In addition, RT-PCR can detect exceedingly rare messages and is also a core method in several subtractive and differential display type techniques that are useful for uncovering differences between two samples. Different primers used in the reverse transcription reaction can be used to highlight different subsets of the RNA population, therefore the methods used might be affected by factors which influence RNA levels.
No previous study has attempted to assess the effect of multiple pre- and postmortem factors on RNA in human postmortem brain tissue. We therefore undertook a multivariate analysis of such factors using three different methods of RT-PCR to measure the housekeeping mRNA, glyceraldehyde 3-phosphate dehydrogenase (GAPdH), in a series of 89 human occipital lobe tissue samples.
2. Materials and Methods
2.1 Tissue selection
Two brain regions of interest were selected on 89 consecutive brains from the Stanley Foundation brain collection maintained under the Cerebral Brain Disorder Branch, Intramural Research Program, National Institute of Mental Health. These brains are collected in a uniform manner by pathologists in selected medical examiners offices in the US. The pathologists are brought to Washington for training to ensure uniform collection methods. The brain regions, taken alternately from the right or left sides of consecutive brains, were as follows:
1. Most lateral aspect of the cerebellar hemisphere at the junction of the inferior and superior semilunar lobe, near the horizontal fissure;
2. Superior-lateral occipital surface of the occipital lobe near the parieto-occipital fissure.
2.2. Rapidity of death measurements
A RoD measure was assigned using the following criteria:
1. Almost instantaneous death, e.g. gunshot wound to the heart, drowning, vehicle accident with death at the scene.
2. Death within 24 h of exciting cause and with minimal evidence of cerebral hypoxia, e.g. vehicle accident with death from internal bleeding occurring several hours later, sedative overdose.
3. Death within 24 h of exciting cause, but with presumption of some cerebral hypoxia, e.g. carbon monoxide poisoning.
4. Slow death occurring over period of >24 h, e.g., death from carcinoma.
5. Slow death with assisted ventilation.
2.3. Freezing and processing of tissue
The brain tissue had been rapidly frozen in 1.5 cm coronal (cerebrum) and horizontal (cerebellum) sections in a slurry of 2-methylbutane (isopentane) (Fluka Chemika, BioChemika, Switzerland) and dry ice and stored at -70 to -75°C for periods from 2 to 24 months. Tissue was trimmed from a frozen wedge of the brain tissue, composed mostly of cortex in the cerebellum and of cortex and underlying white matter in the occipital lobe. Leptomeninges were removed when visible.
2.4. Measurement of pH
A small piece of tissue (~0.5-0.9 g) was homogenized in a polystyrene 10 ml centrifuge tube (with cap) in 10X volume of reverse osmosis deionized water (Hydro Services and Supplies, Durham, NC). The water had been adjusted to pH 7.0 with 0.01 N NaOH before adding to the tissue. Tissue samples were processed by quick submersion of a 7mm diameter disposable generator probe with processing range from 0.25 to 10 ml (Omni International, Gainesville, VA) attached to a hand-held tissue homogenizer (OMNI TH, same vendor) at a variable speed from 5000 to 30,000 rpm (usually midrange) in order to prevent excessive foaming and heating. A single pH value was obtained immediately after each homogenization. Each value was the average of numerous values obtained during an ~ 1 min reading. After each pH measurement, the probe and the electrode were thoroughly rinsed with deionized water. The electrode was cleaned periodically (after each 10 samples) with rinse solution obtained from ATI Orion (vendor as below) in order to prevent clogging of the electrode ceramic reference junction. The accuracy of the electrode was periodically checked using standard buffer at pH 7.00. All tissue homogenates were immediately rapidly frozen in dry ice and stored at -70°C for further use. All procedures were performed at room temperature.
The pH meter (model 370 PerpHect PH/ISE, ATI Analytical Technology, Boston, MA) and the PerpHect ROSS glass semi-micro combination pH electrode (same vendor) were employed in a digital LogR pH measurement mode and manual two-buffer calibration in the range of pH 4.0l and pH 7.00 before each pH measurement was performed. Digital LogR technology uses the electrical resistance of the glass sensing bulb as the temperature probe and is based on the fact that the logarithm of the resistance of the bulb varies almost linearly with the reciprocal of the absolute temperature. This technology allows accurate temperature compensation which is essential for accurate pH measurement.
2.5. RNA purification
Total RNA was purified from 0.5-0.7 g pieces of occipital lobe tissue adjacent to the pieces used for the pH measurement. Tissue was homogenized, with a Brinkman PT 10/35 homogenizer with a PTA-10TS generator, on wet ice in 10 ml Trizol (Gibco BRL) for 2 x 45 s, with 45 s rest, in between, on ice. Two mililitres of chloroform was added and samples were shaken vigorously. They were then transferred to a centrifuge tube and spun at 4°C, 12,000 x g, for 15 min in a Sorvall RC28S centrifuge. The aqueous layer was transferred to a fresh tube and 5 ml isopropanol was added and swirled vigorously to precipitate the RNA. After 10 min at room temperature, the RNA was pelleted by centrifugation at 4ºC, 12,000 x g, for 10 min. The supernatant was decanted and the pellet was washed with 15 ml 70% EtOH. The tubes were spun once more at 4ºC, 12,000 x g, for 10 min. The supernatant was again decanted, the pellet was completely air dried for 15-20 min. and then resuspended in 200 µl DEPC treated water. Concentration of RNA was quantitated by A260.
2.6.DNAse treatment
50 µg of total RNA was brought to 80 µl in DEPC water. The mixture was brought to 100 µl and a final concentration of 40 mM Tris pH 8.0, 10 mM NaCl, 6 mM NgCl2 and 10 mM CaCl2 and 0.2 U/µl RQ1 RNAse free DNAse (Promega). The reactions were incubated for 15 min at 37°C. Reactions were stopped with 4 µl of 0.5 M EDTA pH 8.0. An equal volume of phenol:chloroform:isoamyl alcohol (Gibco BRL) was added to each tube. Tubes were vortexed briefly, then spun 1-2 min in a tabletop microfuge. The aqueous layer was transferred to a fresh tube and 15 µl of 3 M NaOAc, pH 5.4 was added and well mixed in. 250 µl 100% EtOH was added to each tube and mixed by inversion. Tubes were chilled at -80°C overnight, then spun for 1 h at 4°C at top speed in a tabletop microfuge. The supernatant was removed and the pellets air-dried. Pellets were resuspended in 50 µl DEPC treated water and the RNA concentration determined by A260.
2.7. Reverse transcription
Total RNA was reverse transcribed to first strand cDNA. 2.5 µl of DNAsed total RNA was brought to 10 µl final volume with DEPC water. Added to this was either 1 µl of 50 ng/ml random hexamers (Pharmacia), 1 µl 0.5 µl/µl oligo dT12-18(Gibco) or 1 µl 25 ng/ml GAP1241 (see below). The samples were heated to 70ºC for 18 min, then iced for 5 min. Next, 5 µl 5X lst strand buffer (Gibco), 2 µl 0.1 M dithiothreitol (Gibco) and 1 µl 10 mM dNTPs (Pharmacia) was added to the sample. Reactions with random hexamers sat at room temperature for 10 min before proceeding. All samples were then heated to 42ºC for 2 min. 1 µl 200 U/ml Superscript II Reverse Transcriptase (Gibco) was added to all samples and reactions were incubated at 42ºC for 50 min. Samples were heated to 70ºC for 10 min then frozen at -80ºC until use. The RT-PCR methods are diagramed in Fig. 2.
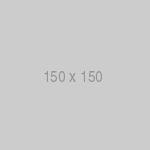
Fig. 2. Reverse transcription and PCR amplification of GAPdH message. Three different primers were used for the reverse transpiration of GAPdH mRNA, as shown above. Oligo dT primers allow transcription from the polyA+ tail region, random hexamers allow transcription all along the mRNA and the GAPdH specific primer (GAPsp) primes directly 5′ of the poly A+ tail region (bases 1260-1241). The amplified PCR product represents the middle third of the 1268 bp message, spanning base pairs 367-894.
2.8. GAPdH PCR
A 1:20 dilution of all first strand cDNAs was made in PCR grade water. One microliter of this dilution was combined with 19 µl water, 2.5 µl each 5 µM GAP primes (see below) and 25 µl PCR Master Mix (Boehringer-Mannheim). A drop of sterile mineral oil (Sigma) was layered on top and the reactions were run on a Hybaid PCR Omnigene (Marsh) thermocycler as follows: 94°C 2 min, (94ºC for 30 sec, 50ºC for 30 sec, 72ºC for 1 min) x 25 cycles, followed by a 10 min extension at 72ºC.
2.9. Visualization and quantitation of the PCR products
The 528 by products of the GAPdH was subjected to polyacrylamide gel electrophoresis on a Hoefer Sturdier gel electrophoresis apparatus. 20 µl of the 50 µl PCR reaction was combined with 4 µl 6X loading buffer (15% Ficoll, 0.25% bromophenol blue), loaded onto a 7.5% polyacrylamide gel (l:37.5 Bis-Acrylamide:Acrylamide, National Diagnostics). 1X in TAE (Gibco) and run at 150 V for 2 h. 0.25 µl of 100 bp ladder (Gibco) were used as standards. Gels were stained for 20 min. in 1:10,000 Syber Green I (Molecular Probes), then visualized on a Molecular Dynamics fluoroimager.
2.10. Quantitation of product
Bands of PCR product were measured on the fluoroimager using ImageQuant software (Molecular Dynamics). The resulting measures, in random fluorescence units, were compared to the demographic data presented in Table 1 in the results section.
2.11. Primer sequence and synthesis
Random hexamers were from Pharmacia, dT12-18 was from Gibco BRL. GAP1241 was synthesized on a Cyclone Plus DNA Synthesizer (Millipore) and a corresponds to bases 1241-1260 on the cDNA sequence (ascension # M33197); 5′-ACA GGG TAC TTT ATT GAT GG-3′.
GAP01 and GAP02 were also synthesized on a Cyclone Plus synthesizer and correspond to bases 367-386 and 894-875, respectively, on the same cDNA sequencer. GAP01; 5′-ACC ACC ATA GAG AAG GCT GG-3′. GAP02; 5′-CTC AGT GTA GCC CAG GAT GC-3′.
2.12. Statistical methods
Logs of the dependent variables were taken to reduce their skewness and improve the explanatory power of the model. Ordinary least squares regression was used (Pindyck and Rubenfeld, 1981). Independent variables (Table 1) were included for cargon monoxide poisoning (CO_DEATH), suicide by other means (OTHSCIDE), mechanical ventilation before death (VENTILTN), tissue storage factors (PMI and FI), RoD (RAPID_2 and RAPID3_5) and occipital pH (OCCIP_PH). SAS software (SAS Institute, 1989) was used.
3. Results
pH measurements were obtained from the occipital and cerebellar lobes of the brains of 89 individuals. Glyceraldehyde phosphate dehydrogenase (GAPdH) mRNA was measured in the occipital lobes by RT-PCR of total RNA.
3.1. ph and demographics
Occipital pH was compared with all demographic data (Table 2). Age ventilation and CO poisoning were all associated with lower values of occipital pH. The strongest correlations were found on the variables representing extreme values of rapidity. A quick onset of death (rapidity=1) was associated with high pH while a rapidity > 3 showed an equally large negative correlation. This supports the theory that pH can be used as a measure of agonal state (Perry et al., 1981; Hardy et al., 1985; Yates et al., 1990). Occipital pH also correlated highly with cerebellar pH (which was measured at the same time. A measure of pH in 10 regions of three different brains (Table 2) shows that pH is remarkably consistent throughout the brain. Thus, any part of the brain may be chosen for use as a predictor of RNA level, even when it is not from the same region used in the RNA preparation.
3.2. GAPdH levels versus pH and demographics
GAPdH mRNA was measured in occipital lobe tissue by RT-PCR. First strand reverse transcription was carried out on total RNA using either oligo dT, random hexamers or a GAP specific primer designed directly 5′ of the polyA tail (Fig. 2). The PCR reaction amplified a region in the middle third of the molecule. By using primers at three different regions, we were able to assess the length of intact RNA downstream of the amplification region.
The three priming methods had different sensitivities to demographic factors as seen in Table 4. FI showed a negative correlation with dT primed reactions. Both random primed and GAPsp reactions had levels that correlate negatively with CO poisoning. This is most likely a secondary effect to the negative correlation between CO poisoning and pH. For all methods of priming, pH was the strongest predictor of cDNA levels. The longer agonal states were regularly associated with the products of all three priming methods, but the strength of the relationships varied between them. While there was a definite and significant trend towards lower message with lower pH, the data were widely scattered (Fig. 3).
Importantly, the levels of GAPdH did not vary by diagnostic group by any method of priming. While all groups showed a wide scatter in the data (Table 5) there was no significant difference by ANOVA analysis. GAPdH levels tended to be lower in depressed individuals for all priming methods. This may be a secondary effect of the high rate of CO deaths in depression (35%) as compared to the other groups (0%, schizophrenic; 19%, bipolar; 10.5%, other psychiatric diagnoses; 0%, no psychiatric diagnoses).
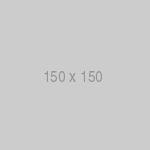
Fig. 3: GAPdh in oligo dT primed cDNA vs. pH. GAPdH levels were measured from oligo dT primed occipital cDNA from 89 brains by RT-PCR as described in Section 2. Occipital pH was measured on the same 89 brains samples, also described in Section 2. The chart below shows the levels of GAPdH in random fluorescence units (rfu) plotted against occipital pH. The median value is denoted by the middle line, the top and bottom lines note the 95% confidence limits. When grouped according to pH, the GAPdH levels vary significantly (p<0.0001) as analyzed by ANOVA (analysis of variance).
3.3. Multiple regression analysis
The factors which showed a statistical significance (p< 0.05) as well as PMI were entered into a multivariate regression model where the contribution and effect of all variables could be compared (Table 6). The pH remained significant in oligo dT and GAPsp priming. Each full point of increase in pH was associated with a non-significant 7-fold increase in RH primed cDNA (p= 0.064), a 13-fold increase in GAPsp-primed cDNA (p= 0.0079) and a 25-fold boost in oligo dT-primed cDNA (p=0.0015). Oligo dT priming also showed the most sensitivity to other variables, with suicide reaching statistical significance and FI narrowly missing significance. Oligo dT-primed cDNA levels were 61% lower in the brains of subjects who had committed suicide (p=0.017) and frozen storage resulted in an average decrease of 7% per month (p=0.071).
4. Discussion
In our RT-PCR analysis of GAPdH levels in postmortem human brains we found that there was no correlation between RNA levels and demographic measures studied such as age, gender, and psychiatric diagnosis. We found that the pH correlated well with RNA as measured by RT-PCR. Northerns, in situs and immunohistochemical blots have been previously shown to correlate to pH (Harrison et al., 1995; Kingsbury et al., 1995) and the relationship appears to remain true even with a very different method of RNA detection. The pH itself correlated best with the RoD scale, making it likely to be strongly influenced by premortem factors. A similar correlation between agonal state and pH was noted in several studies (Perry et al., 1981; Hardy et al, 1985; Yates et al., 1990). While we found that pH works well as a rough measure of RNA levels, there is a great degree of scatter in that data, such that there is no absolute cutoff over which the yield of RNA may be guaranteed.
The pH measurements made in this study are lower than those reported by the Harrison and Kingsbury groups (Harrison et al., 1995; Kingsbury et al., 1995). While the absolute numbers are lower, the same trends are seen in all studies. We are unsure of the reason for this disparity since all three studies share the same technical methods. We remain confident that within this set of data, the correlations between pH and RNA levels are valid.
While the levels of GAPdH vary between samples, when surveyed in large numbers it can be seen that the message does not vary between diagnostic groups. As we look for disease specific variations in message levels, this gives us more confidence that differences between groups are real and not due to qualitative differences in the RNA. A similar finding was reported in translation assays from postmortem samples of affected and unaffected brains (Whatley et. al., 1996). This also underscores the importance of using large sample sets, thus ensuring the averaging out of individual variations.
Our studies indicate RNA levels, as measured by any form of RT-PCR priming, are consistently correlated with premortem factors as reflected by pH. On the postmortem factor of freezer storage, there was a difference between the different priming methods used, apparently due to differential loss of the polyA tail. The loss of polyA tails over an extended FI had been noted previously by Leonard et al. (Leonard et al., 1993) and others had noted their general instability (Octave et al., 1988; Johnson et al., 1986). The loss of the tail does not necessarily lead to the loss of the message as shown by the data for GAPdH specific and random hexamer primed reactions. It is usually thought that the loss of the polyA tail leads to the rapid degradation of the rest of the message (Bernstein et al., 1989). It is possible that damage or loss during storage at -80°C may indeed impair the ability to prime with oligo dT, but that the enzymes and mechanisms responsible for the ensuing breakdown might not be active as such a low temperature. Such a scenario might explain the instability or RNA from thawed tissues reported by (Morrison et al., 1987; Ragsdale and Miledi, 1991), since the thawed enzymes might make short work of the already destabilized poly A-RNA. Caution is therefore advised when using oligo dT priming for comparison between samples that have widely different freezer storage intervals.
Some minor correlations emerged in the course of the multivariate analysis. There was a relationship between suicide and levels of GAPdH in oligo dT primed reactions. Since suicide is a very complex factor it is difficult to speculate on a cause for this correlation. The association found in this study is statistically very marginal, but it is worthy of concern since suicides are by definition excluded from mentally normal control groups. Analysis of a suicide free control group with a group of mentally ill cases with a high rate of suicide could present an artifactual difference between the diagnoses that is actually due to suicide.
We have found it possible to study RNA, from a large number of postmortem human brains using RT-PCR based methods. We were not able to account for the variability between samples, but due to the correlations with pH and RoD, we speculate that they are due to a complex set of mostly premortem factors. By using multivariate methods it is possible to identify a variety of potentially important covariates on RNA product levels that can help researchers to devise appropriate tissue collection methods and analysis plans.
Acknowledgements
The authors would like to thank Dr. Christopher Ross and Dr. Paul Harrison for helpful discussions and advice. Ms. Ann Cusic and Ms. Melissa Skolasky for technical assistance and the members of the Stanley Neuropathology Consortium which includes Joel E. Kleinman, M.D., Ph.D., Thomas M. Hyde, M.D., Ph.D., Llewellyn B. Bigelow, M.D., and Maree J. Webster, Ph.D.
Table 1
Independent variables; averages and ranges of values
| Minimum Value | Maximum Value | Mean | |
| Diagnosis Schizophrenia (N=26, range 23-83 years, median 34.0) Bipolar disorder (N=21,range 21-57 years, median 37.0)Depression (N=13, range 21-65 years, median 40.5) Other diagnoses (N=20, range 17-68 years, median 36.0) Normal (N=9, range 28-59 years, median 48.0) Gender (female, N=26) |
0.00 0.00 0.00 0.00 0.00 0.00 |
1.00 1.00 1.00 1.00 1.00 1.00 |
0.29 0.24 0.15 0.22 0.10 0.29 |
| Cause of death Other suicide (N=32) CO poisoning (N-12) Mechanical ventilation (N=10) Age (years) |
0.00 0.00 0.00 17.00 |
1.00 1.00 1.00 83.00 |
0.36 0.13 0.11 39.66 |
| Rapidity of death 1 (fast) (N=22) 2 (N=25) 3-5 (slow) (N=22) |
0.00 0.00 0.00 |
1.00 1.00 1.00 |
0.47 0.28 0.25 |
| Tissue storage time RI (h) PMI (h) FI (months) |
1.00 5.00 2.00 |
40.0 133.00 24.00 |
7.61 40.87 10.29 |
| pH Cerebellar pH Occipital pH |
5.40 5.50 |
6.30 6.30 |
5.87 5.85 |
For variables that are either true or false for a given sample, the variable is assigned a value of 0.00 if false and 1.00 if true. For other variables, the appropriate units are listed.
TABLE 2
Univariate analysis of brain pH vs. demographic factors
Factors Occipital pH Schizophrenia R= -0.14 Bipolar disease 0.059 Depression -0.15 Other diagnosis 0.068 Normal 0.21 Female -0.13 Other suicide 0.17 CO poisoning -0.23* Ventilation -0.24* Age -0.26* Freezer interval 0.014 Refrigerator interval -0.031 Postmortem interval 0.20 Rapidity = 1 0.29** Rapidity = 2 -0.02 Rapidity = 3-5 -0.32** Occipital pH 1.00 Cerebellar pH 0.70***
Correlation and significant values. pH was measured on samples from the occiptial and cerebellar lobes as described in Section 2. Ordinary least squares regression were run to compare pH against the demographics variables above. *=< 0.05; **p<0.01; *** p< 0.0001.
TABLE 3
Intrabrain pH in 10 regions of three different brains
| Region | ph Brain Aa | pH Brain Bb | pH Brain Cc |
| Frontal | 6.5 | 5.8 | 6.2 |
| Temporal | 6.7 | 5.9 | 6.3 |
| Parietal | 6.5 | 5.8 | 6.4 |
| Occipital | 6.7 | 5.8 | 6.4 |
| Cerebellum | 6.4 | 5.7 | 6.2 |
| Hippocampus | 6.5 | 5.7 | 6.7 |
| Thalamus | 6.4 | 5.7 | 6.3 |
| Caudate | 6.6 | 5.7 | 6.2 |
| Pons | 6.6 | 5.8 | 6.0 |
| Medulla | 6.5 | 6.1 | 6.1 |
| Average | 6.54 | 5.80 | 6.28 |
pH was measured on 10 regions of three different brains in Section 2. Brains were selected for having varying pr- and postmortem factors that wre likely to affect pH.
aBrain A: PMI = 36 h; Rod = 1h; RI = 1 h
bBrain B:PMI = 51 h; Rod = 3 h; RI = 3 h
cBrain C: PMI = 133 h;Rod = 4 h; RI = 4 h
Table 4
Univariate analysis of GAPdH message versus demographic factors
| Factors | Oligo dT primed cDNA | GAPdH specific primed cDNA | Random hexamer primed cDNA |
| Schizophrenia Bipolar disease Depression Other diagnosis Normal Female Other suicide CO Poisoning Ventilation Age Freezer interval Refrigerator interval Postmortem interval Rapidity = 1 Rapidity = 2 Rapidity = 3-5 Occipital pH Cerebellar pH |
R= -0.0081 0.049 -0.14 -0.017 0.12 -0.069 -0.13 -0.19 -0.15 0.085 -0.31** -0.15 -0.20 0.24* -0.026 -0.25* 0.35** 0.27** |
0.0045 0.0023 -0.16 0.10 0.043 -0035 -0.013 -0.26* -0.090 0.070 -0.19 -0.13 -0.16 0.27** -0.12 -0.18 0.34*** 0.28** |
0.048 0.079 -0.19 -0.019 0.062 0.084 -0.071 -0.26* -0.14 0.15 0.098 -0.066 -0.059 0.14 0.089 -0.26* 0.25* 0.36** |
Correlation and significance values. Ordinary least squares regressions were run on the natural logs of the GAPdH levels as described in Section 2. * p< 0.05; **<0.01; ***<0.001.
Table 5
Comparison of GAPdH levels between diagnostic groups
| Diagnosis | Mean | Median | Range |
| Oligo dT primed cDNA Schizophrenia Bipolar Disorder Depression Other psychiatric diagnoses No psychiatric diagnoses |
66,803 68,665 34,439 65,974 85,210 |
11,978 18,176 3,938 12,177 64,549 |
1000-321,097 1000-359,536 1000-246,093 1000-315,600 1000-249-529 |
| GAPdH specific primed cDNA Schizophrenia Bipolar Disorder Depression Other psychiatric diagnoses No psychiatric diagnoses |
40,649 38,787 26,527 44,559 40,371 |
11,383 11,178 4,285 23,437 13,190 |
1000-212,404 1000-196,976 1000-135,793 1000-170,467 1000-130,165 |
| Random hexamer primed cDNA Schizophrenia Bipolar Disorder Depression Other psychiatric diagnoses No psychiatric diagnoses |
87,801 87,295 63,533 78,146 81,810 |
26,566 48,900 11,425 25,995 56,672 |
1586-623,299 100-400,871 581-226,081 1000-319,033 1347-248,943 |
GAPdH was measured by RT-PCR as described in Section 1. The levels above are listed in random fluorescent units.
Table 6
Multivariate analysis of factors affecting GAPdH levels
| Parameter estimates | |||||
| Variable | Parameter estimates | S.E. | T for H0:Parameter=0d | Prob>|T|e | eßf |
| A. Oligo dT primedg F8.83=4.83a (p=0.0002) Intercept CO death Other suicide Rapidity 2 Rapidity 3-5 Ventilation Freezer PMI Occipital pH |
R-square=0.30b -7.38 -1.03 -0.93 -0.16 -0.054 -0.52 -0.076 -0.0090 3.21 |
5.78 |
Adj. R-Sq=0.23c -1.28 -1.14 -2.43 -0.363 -0.071 -0.69 -1.83 -1.123 3.28 |
0.21 |
0.36 |
| B. GAPdH specific primedh F8.83=3.34a(p=0.0024) Intercept CO death Other suicide Rapidity 2 Rapidity 3-5 Ventilation Freezer PMI Occipital pH |
R-Square=0.25b -4.41 -2.22 -0.48 -0.56 0.87 -0.98 -0.043 -0.0052 2.56 |
5.55 |
Adj. R-sq=0.17c -0.80 -2.55 -1.30 -1.33 1.20 -1.34 -1.074 -0.68 2.72 |
0.43 0.013 0.20 0.19 0.23 0.18 0.29 0.50 0.0079 |
0.11 0.57 2.39 0.38 0.96 0.99 12.98 |
| C.Random Hexamerprime F8.83=1.84a (p=0.082) Intercept CO death Other suicide Rapidity 2 Rapidity 3-5 Ventilation Freezer PMI Occipital pH |
R-square=0.15b -1.26 -1.18 -0.20 0.17 0.075 -0.65 0.069 -0.012 1.94 |
6.10 |
Adj R-sq=0.70c -0.21 -1.23 0.49 0.35 0.094 -0.81 1.57 -1.39 1.88 |
0.84 |
0.31 |
Variables for the multivariate model were chosen as described in Section 2. Boldface indicates variables with significant correlations.
aF statistic, tests hypothesis that all parameters equal zero except the intercept, calculated as mean square for model dividen by mean square for error. Subscripts indicate degrees of freedom derived from the model (8) and from the error (83). Prob>F, the probability of getting a greater F statistic than that observed if the hypothesis is true.
bR-square, the proportion of variation in the data that is explained by the model.
cAdj. R-Sq., the adjusted R-square, adjusted for the number of variables.
dT for HO, parameter=O= the t test that parameter is zero, calculated as the parameter estimate divided by the S.E.
eProb>|T|, two-tailed significance.
feß, the effect of the variable on y value as a ratio.
gA, The model applied to RNA levels as measured from RT-PCR of oligo dT primed total RNA.
hB, RNA levels as measured from RT-PCR of GAPdH specific primed total RNA.
iC, RNA levels as measured from RT-PCR of rnadom primed total RNA.
REFERENCES
Barton AJL, Pearson RCA, Najilerahim A, Harrison PJ. Pre- and postmortem influences on brain mRNA. J Neurochem 1993;61:1-11.
Bernstein P, Peltz SW, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol 1989;9:659-70.
Bowen DM, Smith CB, White P, Davidson AN. Neurotransmitter-related enzymes and indices of hypoxia in senile dementia and other abiotrophies. Brain 1976;99:459-96.
Butterworth J, Yates CM, Simpson J. Phosphate activated glutaminase in relatin to Huntington’s disease and agonal state. J neurochem 1983;41:440-7.
Butterworth J. Changes in nine enzyme markers for neurons, glia and endothelial cells in agonal state and Huntington’s disease caudate nucleus. J. Neurochem 1986;47:583-7.
Czudek C, Reynolds GP. 3H-nipecotic acid binding to g-aminobutyric acid uptake sites in post-mortem human brain. J Neurochem 1990;55:165-8.
Dodd PR, Hambley JW, Cowburn RF, Hardy JA. A comparison of methodologies for the study of functional transmitter neurochemistry in human brain. J Neurochem 1988;50:1333-45.
Hardy JA, Wester P, Winblad B, Gezelius C, Bring G, Eriksson A. The patients dying after long terminal phase have acidotic brains; implications for biochemical measurements on autopsy tissue. J Neural Tranm 1985;61:253-64.
Harrison PJ, Procter AW, Barton AJL, Lowe SL, Najlerahim A, Bertolucci PHF, Bowen DM, Pearson RCA. Terminal coma affects messenger RNA detection in post mortem human temporal cortex. Mol Brain Res 1991;9:161-4.
Harrison PJ, Heath PR, Eastwood SL, Burnet PWJ, McDonald B, Pearson RCA. The relative importance of premortem acidosis and postmortem interval for human brain gene expression studies: selective mRNA vunerability and comparison with their encoded proteins. Neurosci Lett 1995;200:151-4.
Johnston SA, Morgan DG, Finch CE. Extensive postmortem stability of RNA from rat and human brain. J Neurosci Res 1986;16:267-80.
Kingsbury AE, Foster OJF, Nisbet AP, Cairns N, Bray L, Eve DJ, Lees AJ, Marsden CD. Tissue pH as an indicator of mRNA preservation in human post-mortem brain. Mol Brain Res 1995;28:311-8.
Leonard S, Logel J, Luthman D, Casanova M, Kirch D, Freedman R. Biological stability of mRNA isolated from human post-mortem brain collection. Biol Psych 1993;33:456-66.
Morrison MR, Pardue S, Maschoff K, Griffin WST, White III, CL, Gilbert J, Roses A. Brain messenger RNA levels and ribonuclease activity in Alzheimer’s disease. Biochem Soc Trans 1987;15:133-4.
Octave J-N, de Sauvage F, Macq A-F, Maloteaux JM. Cloning of the cDNA from normal brain and brain of patients with Alzheimer’s disease in the expression vector lambda gt11. Prog Neuro-Psychopharmacol Biol Psychiatry 1988;12:813-20.
Perrett CW, Marchbanks RM, Whatley SA. Characterisation of messenger RNA extracted post-mortem from the brains of schizophrenic, depressed and control subjects. J Neurol Neurosurg Psychiatry 1988;51:325-31.
Perry EK, Perry RH, Tomlinson BE. The influence of agonal states on some neurochemical activities of postmortem human brain tissue. Neurosci Lett 1981;29:303-7.
Pindyck RS, Rubinfeld DL. Econometric Models and Economic Forecasts. 2nd ed. New York:McGraw-Hill, 1981:630.
Ragsdale DS, Miledi R. Expressional potency of mRNAs encoding receptors and voltage-activated channels in the postmortem rat brain. Proc Natl Acad Sci USA 1991:88:1854-8.
Spokes EGS. An analysis of factors influencing measurements of dopamine, noradrenaline, glutamate decarboxylase and choline acetylase in human postmortem brain tissue. Brain 1979;102:333-46.
Taylor GR, Carter GI, Crow TJ, Johnson JA, Fairbairn AF, Perry EK, Perry RH. Recovery and measurement of RNA in Alzheimer’s disease by molecular hybridization. J Neurol Neurosurg Psychiatry 1986;50:336.
Whatley SA, Curti D, Marchbanks RM. Mitochondrial involvement in schizophrenia and other functional psychoses. Neurochem Res 1996;21:995-1004.
Yates CM, Butterworth J, Tennant MC, Gordon A. Enzyme activities in relation to pH and lactate in postmortem brain in Alzheimer’s-type and other dementias. J. Neurochem 1990;55:1624-30.
